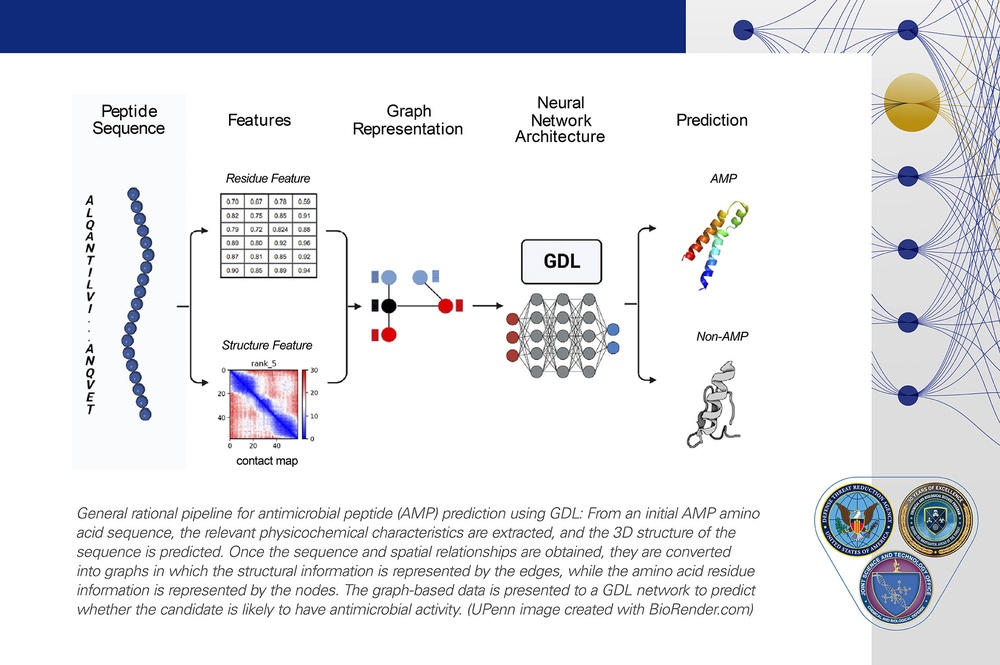
General rational pipeline for antimicrobial peptide (AMP) prediction using GDL: From an initial AMP amino acid sequence, the relevant physicochemical characteristics are extracted, and the 3D structure of the sequence is predicted. Once the sequence and spatial relationships are obtained, they are converted into graphs in which the structural information is represented by the edges, while the amino acid residue information is represented by the nodes. The graph-based data is presented to a GDL network to predict whether the candidate is likely to have antimicrobial activity. (UPenn image created with BioRender.com)
| Date Taken: | 08.12.2024 |
| Date Posted: | 08.12.2024 22:58 |
| Photo ID: | 8586141 |
| VIRIN: | 240812-D-D0490-1005 |
| Resolution: | 2083x1385 |
| Size: | 429.52 KB |
| Location: | FT. BELVOIR, VIRGINIA, US |
| Web Views: | 81 |
| Downloads: | 1 |

This work, From Years to Hours: Accelerating Drug Discovery with Advanced ML Techniques, must comply with the restrictions shown on https://www.dvidshub.net/about/copyright.